Back to Courses





Health Informatics Courses - Page 8
Showing results 71-80 of 93

Social Determinants of Health: Health Care Systems
This third of five courses explores topics related to the social determinants of health and health care systems. This course will also focus on the relationship between the social determinants of health, mental health, substance abuse, and trauma. The topics of this course include:
1. Health Literacy
2. Mental Health & Substance Abuse
3. Violence, Conflict, and Trauma
4. Ethical Considerations for Health Systems and Data
5. Data Applications: Correlation Analysis and Heat Map Visualization

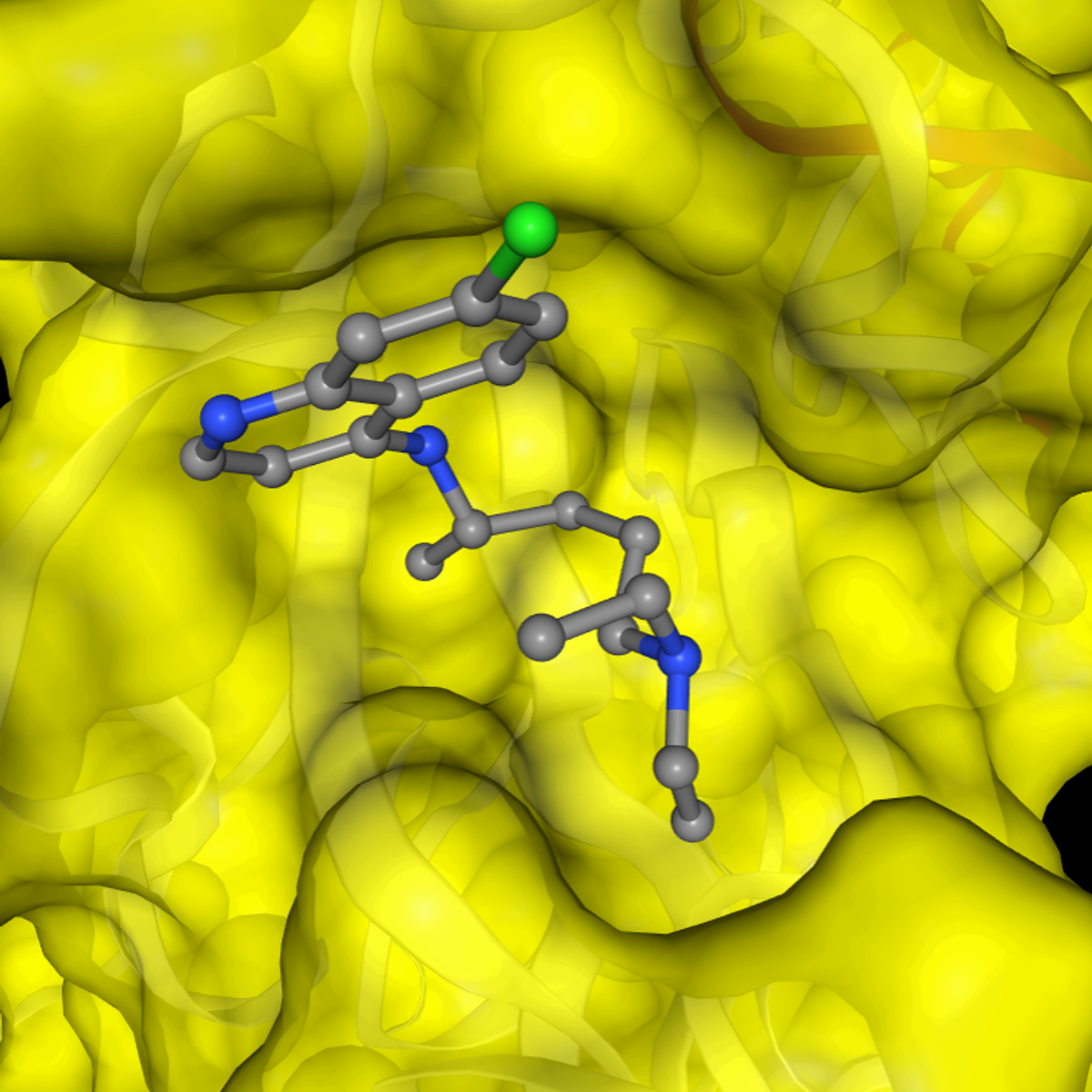
SARS-CoV-2 Protein Modeling and Drug Docking
In this 1-hour long project-based course, you will construct a 3D structure of a SARS-CoV-2 protein sequence using homology modeling and perform molecular docking of drugs against this protein molecule and infer protein-drug interaction. We will accomplish it in by completing each task in the project which includes
- Model protein structures from sequence data
- Process proteins and ligands for docking procedure
- Molecular docking of drugs against protein molecules
Note: This course works best for learners who are based in the North America region. We’re currently working on providing the same experience in other regions.

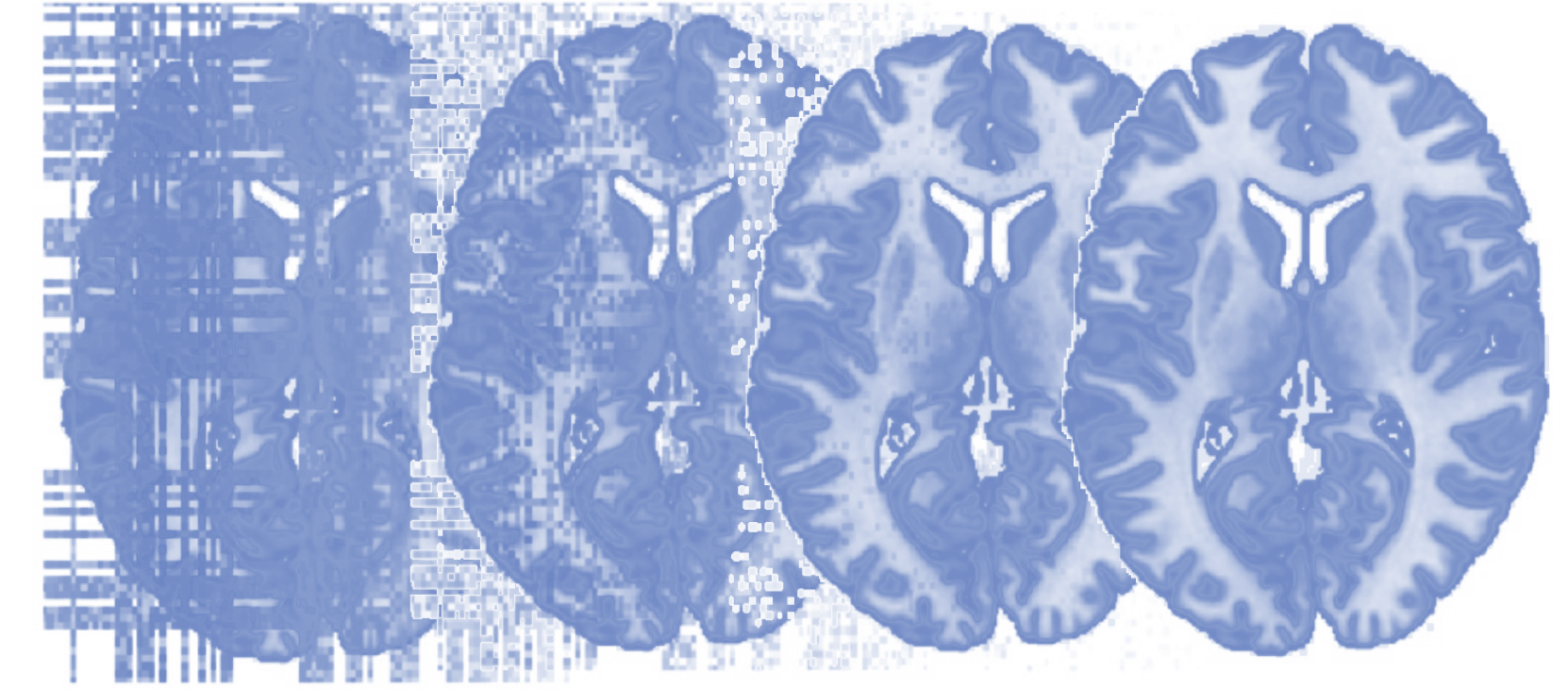
Principles of fMRI 1
Functional Magnetic Resonance Imaging (fMRI) is the most widely used technique for investigating the living, functioning human brain as people perform tasks and experience mental states. It is a convergence point for multidisciplinary work from many disciplines. Psychologists, statisticians, physicists, computer scientists, neuroscientists, medical researchers, behavioral scientists, engineers, public health researchers, biologists, and others are coming together to advance our understanding of the human mind and brain. This course covers the design, acquisition, and analysis of Functional Magnetic Resonance Imaging (fMRI) data, including psychological inference, MR Physics, K Space, experimental design, pre-processing of fMRI data, as well as Generalized Linear Models (GLM’s). A book related to the class can be found here: https://leanpub.com/principlesoffmri.

Fundamentals of Machine Learning for Healthcare
Machine learning and artificial intelligence hold the potential to transform healthcare and open up a world of incredible promise. But we will never realize the potential of these technologies unless all stakeholders have basic competencies in both healthcare and machine learning concepts and principles.
This course will introduce the fundamental concepts and principles of machine learning as it applies to medicine and healthcare. We will explore machine learning approaches, medical use cases, metrics unique to healthcare, as well as best practices for designing, building, and evaluating machine learning applications in healthcare.
The course will empower those with non-engineering backgrounds in healthcare, health policy, pharmaceutical development, as well as data science with the knowledge to critically evaluate and use these technologies.
Co-author: Geoffrey Angus
Contributing Editors:
Mars Huang
Jin Long
Shannon Crawford
Oge Marques
The Stanford University School of Medicine is accredited by the Accreditation Council for Continuing Medical Education (ACCME) to provide continuing medical education for physicians. Visit the FAQs below for important information regarding 1) Date of original release and Termination or expiration date; 2) Accreditation and Credit Designation statements; 3) Disclosure of financial relationships for every person in control of activity content.

Introduction to Digital Health Entrepreneurship
This course will provide an overview of digital health entrepreneurship with an initial emphasis on learning the basic digital health terminology, exploring a current example of telemedicine and its growth during the COVID pandemic, and gaining an understanding of the landscape and macro forces that affect the US healthcare system and the evolution of digital health. Students who successfully complete the course will understand the impact that payers, regulators, clinicians, and patients have on and the implications for digital health innovations.

Genome Sequencing (Bioinformatics II)
You may have heard a lot about genome sequencing and its potential to usher in an era of personalized medicine, but what does it mean to sequence a genome?
Biologists still cannot read the nucleotides of an entire genome as you would read a book from beginning to end. However, they can read short pieces of DNA. In this course, we will see how graph theory can be used to assemble genomes from these short pieces. We will further learn about brute force algorithms and apply them to sequencing mini-proteins called antibiotics.
In the first half of the course, we will see that biologists cannot read the 3 billion nucleotides of a human genome as you would read a book from beginning to end. However, they can read shorter fragments of DNA. In this course, we will see how graph theory can be used to assemble genomes from these short pieces in what amounts to the largest jigsaw puzzle ever put together.
In the second half of the course, we will discuss antibiotics, a topic of great relevance as antimicrobial-resistant bacteria like MRSA are on the rise. You know antibiotics as drugs, but on the molecular level they are short mini-proteins that have been engineered by bacteria to kill their enemies. Determining the sequence of amino acids making up one of these antibiotics is an important research problem, and one that is similar to that of sequencing a genome by assembling tiny fragments of DNA. We will see how brute force algorithms that try every possible solution are able to identify naturally occurring antibiotics so that they can be synthesized in a lab.
Finally, you will learn how to apply popular bioinformatics software tools to sequence the genome of a deadly Staphylococcus bacterium that has acquired antibiotics resistance.
Data mining of Clinical Databases - CDSS 1
This course will introduce MIMIC-III, which is the largest publicly Electronic Health Record (EHR) database available to benchmark machine learning algorithms. In particular, you will learn about the design of this relational database, what tools are available to query, extract and visualise descriptive analytics.
The schema and International Classification of Diseases coding is important to understand how to map research questions to data and how to extract key clinical outcomes in order to develop clinically useful machine learning algorithms.

Trustworthy AI for Healthcare Management
This MOOC gives an introduction to trustworthy artificial intelligence and its application in healthcare. This includes modules on basics of artificial intelligence and an introduction to trustworthy and ethical applications of artificial intelligence. A dedicated lesson will present the Z-Inspection® process for assessing trustworthy AI, and real-world case studies will illustrate how to apply the knowledge.
The course is aimed at healthcare professionals, patients, and AI practitioners. It does not require previous knowledge on AI.
Participants will learn:
- How AI systems work and learn
- What tasks can be solved by AI
- Common challenges for AI in healthcare
- Fundamentals of Trustworthy AI
- How to assess Trustworthy AI
- How Trustworthy AI assessments look in practice
This course was developed by Goethe University Frankfurt as part of the research project 'Pan-European Response to the Impacts of the COVID-19 and future Pandemics and Epidemics' (PERISCOPE). Funded by the European Commission Research Funding programme Horizon 2020 under the Grant Agreement number 101016233, PERISCOPE investigates the broad socio-economic and behavioural impacts of the COVID-19 pandemic, to make Europe more resilient and prepared for future large-scale risks.
PERISCOPE website: https://www.periscopeproject.eu/

Hacking COVID-19 — Course 3: Unraveling COVID-19's Origins
In this course, you will follow in the footsteps of the bioinformaticians investigating the COVID-19 outbreak by investigating the origins of SARS-CoV-2. Whether you’re new to the world of computational biology, or you’re a bioinformatics expert seeking to learn about its applications in the COVID-19 pandemic, or somewhere in between, this course is for you! As you go through this journey, we will introduce and explain genomic concepts and give you many opportunities to practice your skills, and we will provide a series of problems with gradually increasing complexity. This third course will only discuss the multiple sequence alignment and maximum-likelihood phylogenetic inference of SARS-CoV-2 genomes, but future courses in this series will explore follow-up bioinformatics analyses used in the COVID-19 pandemic.

Foundations of Telehealth
Telemedicine has proven itself to be an important part of the future of healthcare. In this course, students will be introduced to the key components and considerations needed to design and implement a successful telemedicine program at both the practice and health system levels. The course emphasizes operational design principles and highlights a team based approach. Key content areas include clinical considerations, patient safety, technology needs, patient satisfaction, legal, government affairs, regulatory and compliance, and billing considerations.
Popular Internships and Jobs by Categories
Find Jobs & Internships
Browse
© 2024 BoostGrad | All rights reserved